A new way to identify bacteria in fluids
A laser can be shined on a drop, of blood, or mucus. The light reflecting back from the laser can be used to identify bacteria in that sample.

A modified version of the Stanford University logo, printed using droplets that contain a 1:1 mixture Staphylococcus epidermidis On a slide coated with gold, bacteria and red blood cells from mice (RBCs), were printed. Droplets were printed using an acoustic transmitter at 147 MHz. (Image credit: Fareeha Safir)
“We can find out not just that bacteria are present, but specifically which bacteria are in the sample – E. coli, Staphylococcus, Streptococcus, Salmonella, anthrax, and more,” said Jennifer Dionne, an associate professor of materials science and engineering and, by courtesy, of radiology at Stanford University. “Every microbe has its own unique optical fingerprint. It’s like the genetic and proteomic code scribbled in light.”
Dionne is senior editor of a new journal study Nano Letters She explains an innovative method her team invented that could allow for faster, cheaper and more accurate microbial tests of almost any fluid.
It can take several hours, if not days, to use traditional culturing techniques. A tuberculosis culture takes 40 days, Dionne said. This new test can be completed in minutes. The team claims that it will provide faster and better diagnosis of tuberculosis, safer antibiotic use, improved environmental monitoring and quicker drug development.
Old dogs are capable of learning new tricks
It is not the discovery that bacteria can display these spectral fingerprints (a fact that has long been known), but the way the team was able to uncover those spectra among the blinding arrays of light that each sample reflects.
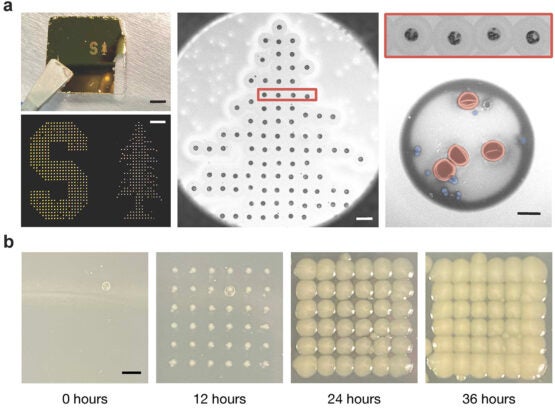
Detail of the gold-coated slide’s printed dots (a). The false coloring of one dot in this close-up shows red blood cells in red. Staphylococcus epidermidis Blue bacteria. To illustrate how the dots react to incubation, the researchers also printed onto an absorbent slide (b). (Image credit: Fareeha Safir)
“Not only does each type of bacterium demonstrate unique patterns of light but virtually every other molecule or cell in a given sample does too,” said first author Fareeha Safir, a PhD student in Dionne’s lab. “Red blood cells, white blood cells, and other components in the sample are sending back their own signals, making it hard if not impossible to distinguish the microbial patterns from the noise of other cells.”
A milliliter of blood – about the size of a raindrop – can contain billions of cells, only a few of which might be microbes. The team needed to isolate and amplify the light reflected from bacteria. To do that, they ventured along several surprising scientific tangents, combining a four-decade-old technology borrowed from computing – the inkjet printer – and two cutting-edge technologies of our time – nanoparticles and artificial intelligence.
“The key to separating bacterial spectra from other signals is to isolate the cells in extremely small samples. We use the principles of inkjet printing to print thousands of tiny dots of blood instead of interrogating a single large sample,” explained co-author Butrus “Pierre” Khuri-Yakub, a professor emeritus of electrical engineering at Stanford who helped develop the original inkjet printer in the 1980s.
“But you can’t just get an off-the-shelf inkjet printer and add blood or wastewater,” Safir emphasized. The researchers modified the printer so that biological samples could be printed using acoustic pulses to circumvent the difficulties of handling them. Each dot of printed blood is then just two trillionths of a liter in volume – more than a billion times smaller than a raindrop. These droplets can hold only a few cells, even though they are tiny at that scale.
Researchers also infused the samples with nanorods of gold that attach to bacteria. They act as antennas and draw the laser light towards the bacteria, amplifying its signal by around 1500 times its normal strength. The bacterial spectrum was amplified and isolated in a way that is scientifically useful.
The final piece is machine learning, which allows you to compare the different spectra from each printed dot in fluid and identify any bacteria.
“It’s an innovative solution with the potential for life-saving impact. We are now excited for commercialization opportunities that can help redefine the standard of bacterial detection and single-cell characterization,” said senior co-author Amr Saleh, a former postdoctoral scholar in Dionne’s lab and now a professor at Cairo University.
Catalyst is a platform for collaboration
This kind of collaboration is a hallmark in the Stanford tradition, where experts from seemingly different fields come together to solve longstanding problems that have societal significance.
This particular approach was hatched during a lunchtime meeting at a café on campus and, in 2017, was among the first recipients of a series of $3 million grants distributed by Stanford’s Catalyst for Collaborative Solutions. Catalyst grants were specifically created to inspire interdisciplinary risk taking and collaboration among Stanford researchers working in high-reward fields like security, health care, environment, autonomy, and safety.
Dionne believes that this technique can be applied to other types of fluids and target cells. She has tested it on blood samples and is confident it will work with any type of fluid.
Stanford also has co-authors such as Loza Tadesse (ex-PhD student); Kamyar Firouzi (research staff); Niaz Baei, professor of pathology at the School of Medicine; Stefanie Jeffrey, Emerita John and Marva Warnock Professor in the School of Medicine. Nhat Vu, Pumpkinseed Technologies, is also a coauthor. Stanford Bio-X also includes Jeffrey, Banaei and Khuri-Yakub. Dionne is also senior vice provost for research platforms/shared infrastructures, a member the Cardiovascular Institute, the Wu Tsai Neurosciences Institute and an affiliate to the Precourt Institute for Energy. Jeffrey is also a member the Stanford Cancer Institute. Khuri-Yakub is also a member of the Cardiovascular Institute, the Stanford Cancer Institute and the Wu Tsai Neurosciences Institute.
The Stanford Catalyst for Collaborative Solutions and the Chan Zuckerberg Biohub Investigator Program funded this research. Seed funds from the Stanford Center for Innovation in Global Health were also used to fund the research. Part of this work was performed at the Stanford Nano Shared Facilities (SNSF) and the Soft & Hybrid Materials Facility (SMF), which are supported by the National Science Foundation and National Nanotechnology Coordinated Infrastructure.
Subscribe to the biweekly for all Stanford science stories Stanford Science Digest.


